Halistanol sulfates I and J, new SIRT1-3 inhibitory steroid sulfates from a marine sponge of the genus Halichondria
Nakamura, F., Kudo, N., Tomachi, Y., Nakata, A., Takemoto, M., Ito, A., Tabei, H., Arai, D., de Voogd, N., Yoshida, M., Nakao, Y., Fusetani, N.(2018) J Antibiot (Tokyo) 71: 273-278
- PubMed: 29184120
- DOI: https://doi.org/10.1038/ja.2017.145
- Primary Citation of Related Structures:
5Y4H - PubMed Abstract:
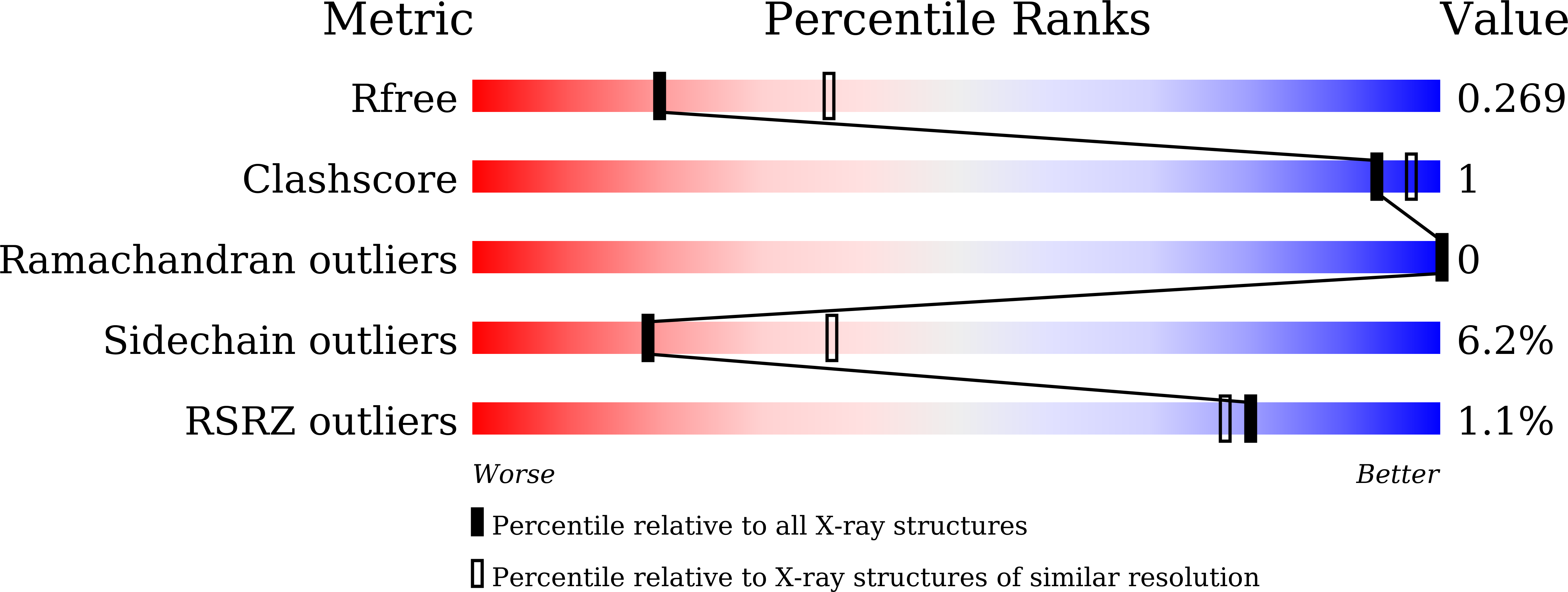
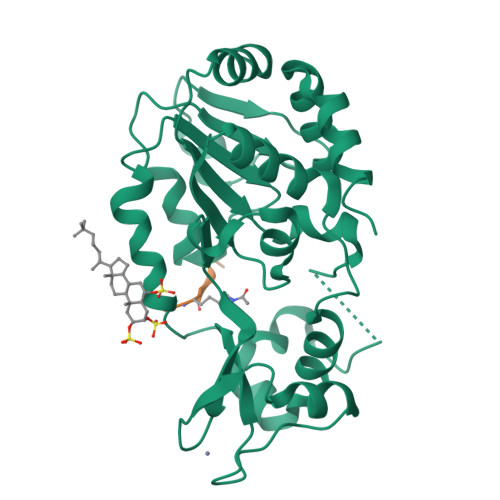
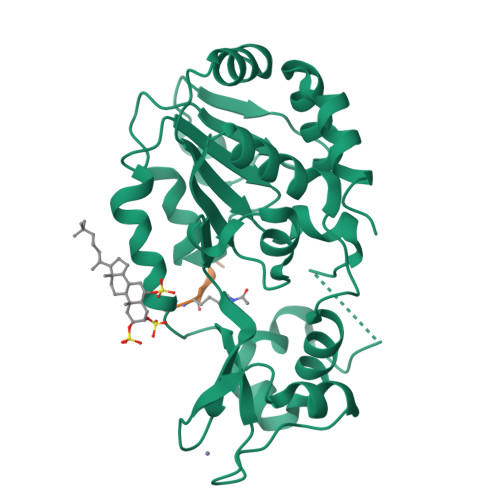
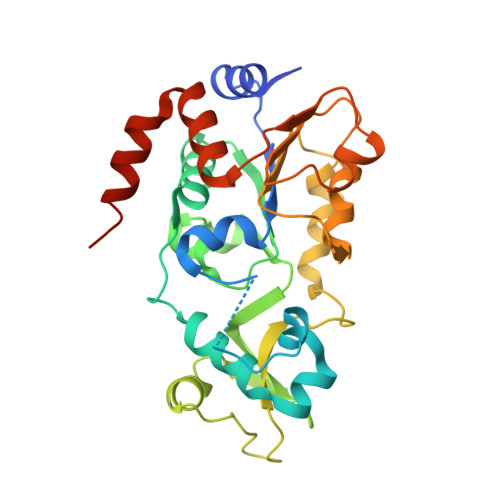
Two new analogs of halistanol sulfate (1) were isolated from a marine sponge Halichondria sp. collected at Hachijo-jima Island. Structures of these new halistanol sulfates I (2) and J (3) were elucidated by spectral analyses. Compounds 1-3 showed inhibitory activity against SIRT 1-3 with IC 50 ranges of 45.9-67.9, 18.9-21.1 and 21.8-37.5 μM, respectively. X-ray crystallography of the halistanol sulfate (1) and SIRT3 complex clearly indicates that 1 binds to the exosite of SIRT3 that we have discovered in this study.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Graduate School of Advanced Science and Engineering, Waseda University, Tokyo, Japan.